Design
The UK National Core Studies—Longitudinal Health and Wellbeing programme (https://www.ucl.ac.uk/covid-19-longitudinal-health-wellbeing/) combines data from multiple UK population-based LS and electronic health records (EHR) to answer pandemic-relevant questions. In this analysis we pooled results from parallel analyses within individual LS, then compared with population-based findings from EHR capturing individuals who actively sought healthcare.
Sample
LS
Data were drawn from 10 UK LS that had conducted surveys before and during the COVID-19 pandemic comprising five age-homogenous cohorts: the Millennium Cohort Study (MCS)28; the Avon Longitudinal Study of Parents and Children (ALSPAC (generation 1, “G1”))29; Next Steps (NS)30; the 1970 British Cohort Study (BCS)31; and the National Child Development Study (NCDS)32, and five age-heterogeneous samples were included: the Born in Bradford study (BIB)33; Understanding Society (USOC)34; Generation Scotland: the Scottish Family Health Study (GS)35; the parents of the ALSPAC-G1 cohort, whom we refer to as ALSPAC-G036; and the UK Adult Twin Registry (TwinsUK)37. Study details and references are shown in Supplementary Table 1. Minimum inclusion criteria were pre-pandemic health measures, age, sex, ethnicity plus self-reported COVID-19, and self-reported duration of COVID-19 symptoms. Ethics statements presented in Supplementary Table 2.
Electronic health records (EHR)
Working on behalf of NHS England, we conducted a population-based cohort study to measure long COVID recording in EHR data from primary care practices using TPP SystmOne software, linked to Secondary Uses Service (SUS) data (containing hospital records) through OpenSAFELY (https://www.opensafely.org/). This is a data analysis platform developed on behalf of NHS England during the COVID-19 pandemic to allow near real-time analysis of pseudonymised primary care records within the EHR vendor’s highly secure data environment to protect patient privacy. Details on Information Governance for the OpenSAFELY platform can be found in the Supplementary Note 1. From a population of all people alive and registered with a general practice on 1 December 2020, we selected all patients who had evidence of a COVID-19-related code, either: positive SARS-CoV-2 testing, being hospitalised with an associated COVID diagnostic code, or having a recorded diagnostic code for COVID in primary care.
Measures
Outcomes: COVID-19 and long COVID definitions
LS: COVID-19 cases were defined by self-report, including testing confirmation and healthcare professional diagnosis (see Supplementary Data 1 for full details of the questions and coding used within each study). Long COVID was defined as per NICE categories using self-reported symptom duration1. Based on these categories, we defined two primary outcomes: (i) symptoms lasting 4+ weeks (symptoms lasting 0–4 weeks as reference) and (ii) symptoms lasting 12+ weeks (symptoms lasting 0–12 weeks as reference). Some studies recorded duration of symptoms of any severity, whereas others referred only to symptoms which impacted daily function (Table 2). In addition, two studies derived alternate estimates of long COVID based on individual symptom counts lasting more than 4 or 12 weeks over at least six months (BiB, TwinsUK) (Supplementary Note 2). All data used to derive these outcomes were collected between April and November 2020.
EHR: Any record of long COVID in the primary care record was coded as a binary variable. This was defined using a list of 15 UK SNOMED codes, categorised as diagnostic (2 codes), referral3 and assessment10 codes. SNOMED is an international structured clinical coding system for use in EHR38. These clinical codes were designed based on guidance issued on long COVID by the NICE1. The outcome was measured between the study start date (1 February 2020) and the end date (9 May 2021).
Exposures
Sociodemographic factors
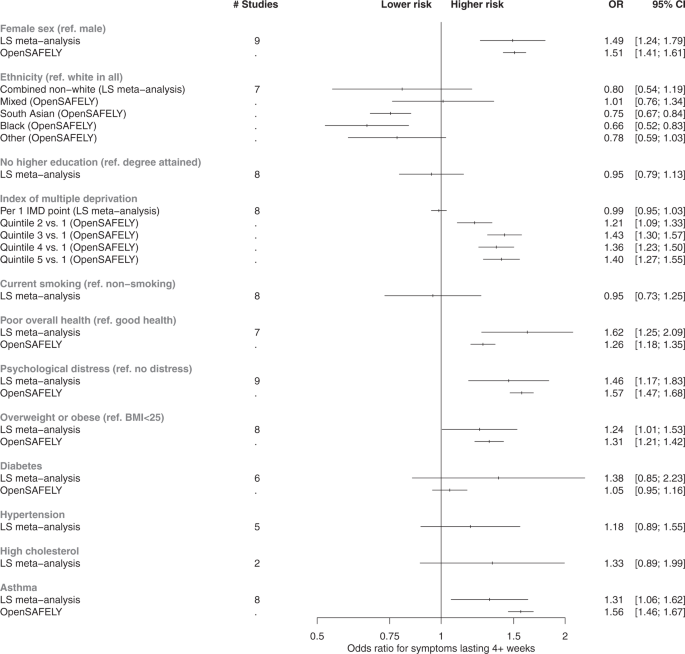
All studies included age, sex, ethnicity (white or non-white minority ethnic group, where available) and Index of Multiple Deprivation (IMD; divided into quintiles with 1 representing the most deprived and 5 representing the least deprived). Area-level SES was measured using the IMD 2019, a composite of different domains including area-level income, employment, education access and crime, for the postcode where a participant lived at the time of sample collection39. LS included additional measures of socioeconomic position: education (degree, no degree), and occupational class of own current/recent employment (Supplementary Data1). EHR also included geographic region40.
Mental health
LS: Pre-pandemic measures using validated continuous scales of anxiety and depression symptoms dichotomised using established cut-offs to indicate distress (see Supplementary Data 1).
EHR: Evidence of a pre-existing mental health condition was defined using prior codes for one of: psychosis; schizophrenia; bipolar disorder; or depression.
Self-rated general health
LS: Pre-pandemic self-rating on a 5-point scale dichotomised to compare excellent-good health (categories 1–3) with fair-poor health (categories 4 and 5).
Overweight and obesity
LS: Body mass index (BMI; kg/m2) obtained prior to the pandemic, coded to compare a BMI between 0 and 24.9 (having underweight/normal weight) against a BMI of ≥25 (overweight/obesity).
EHR: Categorised as having or not having obesity using the most recent BMI measurement, with those having obesity further classified into having Obese I (BMI 30–34.9), Obese II (BMI 35–39.9), or Obese III (BMI 40+). A BMI of >25 was used in LS as the percentage of those in the obese category (i.e., BMI > 30) was relatively small, e.g., 8.9% for TwinsUK, whereas EHR obesity codes were used as these are more reliable and valid indicators of having obesity in general practice.
Health conditions
LS: Pre-pandemic self-report of asthma, diabetes, hypertension, and high cholesterol status.
EHR: A previous code 6 months to 5 years before March 2020 for one or more of: diabetes; cancer; haematological cancer; asthma; chronic respiratory disease; chronic cardiac disease; chronic liver disease; stroke or dementia; other neurological condition; organ transplant; dysplasia; rheumatoid arthritis, systemic lupus erythematosus or psoriasis; or other immunosuppressive conditions. Those with no relevant code for a condition were assumed not to have that condition. Number of conditions were categorised into “0”, “1”, and “2 or more”.
Health behaviours
LS: Current smoking status (dichotomised into “0” = no, “1” = yes).
Statistical analysis: LS
Main analyses were conducted in studies with a direct self-reported measure of COVID-19 symptom length. Associations between each factor and both long COVID outcomes (symptoms for 4+ weeks and symptoms for 12+ weeks) were assessed in separate logistic regression models within each study. We adjust for a minimal set of covariates across all studies, where relevant: age (adjusted as a continuous variable when being considered a covariate), sex, and ethnicity. We report odds ratios (ORs) and 95% confidence intervals (CIs). To synthesise association magnitudes across studies, fixed-effect meta-analysis with restricted maximum likelihood was carried out and repeated with random-effects modelling for comparison. The I2 statistic was used to report heterogeneity between estimates. Meta-analyses were conducted using the metafor package41 for R version 4).
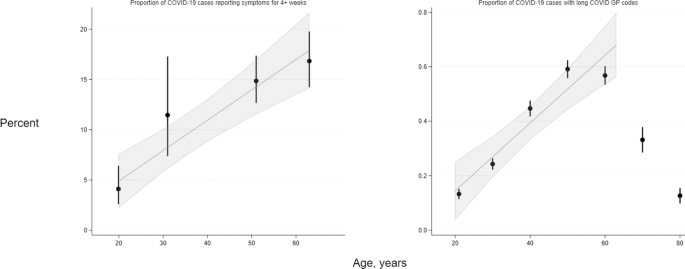
Due to the different age structures of the LS, examination of the direct relationship of age with long COVID risk was treated distinctly from other risk factors, and we modelled the relationship in two ways. First, in age-heterogeneous samples we compared long COVID risk within pre-defined age categories, comparing 45–69 and 70+ to 18–44 in three cohorts (USOC, TwinsUK and GS), and 55–59 and 60–76 to 45–54 in one cohort (ALSPAC G0). Second, in a subset of LS birth cohorts with participants of near-identical ages and who were issued fully harmonised long COVID questionnaires (MCS, NS, BCS70 and NCDS), we analysed the trend in absolute risk of long COVID with increasing age between studies using meta-regression.
Attrition and survey design were addressed by weighting estimates to be representative of their target population in each LS (weights were not available for BiB and TwinsUK).
Sensitivity analyses
To mitigate index event bias27, IPW were derived for risk of COVID-19. These were derived in each LS separately but following a common approach used previously (see Supplementary Note 3 for detail)42. Derived weights were then applied in all analysis models as a sensitivity check.
For studies in which we were able to verify SARS-CoV-2 infection (TwinsUK and ALSPAC-G0 and -G1), analyses were repeated on the sub-sample of those who had positive polymerase chain reaction (PCR) obtained through linkage to testing data and/or lateral flow antibody testing (ALSPAC) and enzyme-linked immunosorbent assay (ELISA) (TwinsUK)43 results confirming viral exposure. These results are presented in Supplementary Figs. 11–14.
Statistical analysis: EHR
We conducted logistic regression to assess whether GP-recorded long COVID was associated with each sociodemographic or pre-pandemic health characteristic. We adjusted for the same set of confounders as used in the LS analyses: age (as categorical variable), sex, ethnicity.
In further analyses of age as a risk factor for long COVID in the EHR data, we assigned individuals within 10-year categories an age at the midpoint of each group, then assessed the trend in long COVID frequency with age using linear and non-linear meta-regression.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.